18. Non-Conjugate Priors#
GPU
This lecture was built using a machine with access to a GPU — although it will also run without one.
Google Colab has a free tier with GPUs that you can access as follows:
Click on the “play” icon top right
Select Colab
Set the runtime environment to include a GPU
!pip install numpyro jax
This lecture is a sequel to the Two Meanings of Probability.
That lecture offers a Bayesian interpretation of probability in a setting in which the likelihood function and the prior distribution over parameters just happened to form a conjugate pair in which
application of Bayes’ Law produces a posterior distribution that has the same functional form as the prior
Having a likelihood and prior that are conjugate can simplify calculation of a posterior, facilitating analytical or nearly analytical calculations.
But in many situations the likelihood and prior need not form a conjugate pair.
after all, a person’s prior is his or her own business and would take a form conjugate to a likelihood only by remote coincidence
In these situations, computing a posterior can become very challenging.
In this lecture, we illustrate how modern Bayesians confront non-conjugate priors by using Monte Carlo techniques that involve
first cleverly forming a Markov chain whose invariant distribution is the posterior distribution we want
simulating the Markov chain until it has converged and then sampling from the invariant distribution to approximate the posterior
We shall illustrate the approach by deploying a powerful Python library, NumPyro that implements this approach.
As usual, we begin by importing some Python code.
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats as st
from typing import NamedTuple, Sequence
import jax.numpy as jnp
from jax import random
import numpyro
from numpyro import distributions as dist
import numpyro.distributions.constraints as constraints
from numpyro.infer import MCMC, NUTS, SVI, Trace_ELBO
from numpyro.optim import Adam
18.1. Unleashing MCMC on a binomial likelihood#
This lecture begins with the binomial example in the Two Meanings of Probability.
That lecture computed a posterior
analytically via choosing the conjugate priors,
This lecture instead computes posteriors
numerically by sampling from the posterior distribution through MCMC methods, and
using a variational inference (VI) approximation.
We use numpyro with assistance from jax to approximate a posterior distribution.
We use several alternative prior distributions.
We compare computed posteriors with ones associated with a conjugate prior as described in Two Meanings of Probability.
18.1.1. Analytical posterior#
Assume that the random variable \(X\sim Binom\left(n,\theta\right)\).
This defines a likelihood function
where \(Y=k\) is an observed data point.
We view \(\theta\) as a random variable for which we assign a prior distribution having density \(f(\theta)\).
We will try alternative priors later, but for now, suppose the prior is distributed as \(\theta\sim Beta\left(\alpha,\beta\right)\), i.e.,
We choose this as our prior for now because we know that a conjugate prior for the binomial likelihood function is a beta distribution.
After observing \(k\) successes among \(N\) sample observations, the posterior probability distribution of \( \theta \) is
Thus,
The analytical posterior for a given conjugate beta prior is coded in the following
def simulate_draw(θ, n):
"""Draws a Bernoulli sample of size n with probability P(Y=1) = θ"""
rand_draw = np.random.rand(n)
draw = (rand_draw < θ).astype(int)
return draw
def analytical_beta_posterior(data, α0, β0):
"""
Computes analytically the posterior distribution
with beta prior parametrized by (α, β)
given # num observations
Parameters
---------
num : int.
the number of observations after which we calculate the posterior
α0, β0 : float.
the parameters for the beta distribution as a prior
Returns
---------
The posterior beta distribution
"""
num = len(data)
up_num = data.sum()
down_num = num - up_num
return st.beta(α0 + up_num, β0 + down_num)
18.1.2. Two ways to approximate posteriors#
Suppose that we don’t have a conjugate prior.
Then we can’t compute posteriors analytically.
Instead, we use computational tools to approximate the posterior distribution for a set of alternative prior distributions using numpyro.
We first use the Markov Chain Monte Carlo (MCMC) algorithm.
We implement the NUTS sampler to sample from the posterior.
In that way we construct a sampling distribution that approximates the posterior.
After doing that we deploy another procedure called Variational Inference (VI).
In particular, we implement Stochastic Variational Inference (SVI) machinery in numpyro.
The MCMC algorithm supposedly generates a more accurate approximation since in principle it directly samples from the posterior distribution.
But it can be computationally expensive, especially when dimension is large.
A VI approach can be cheaper, but it is likely to produce an inferior approximation to the posterior, for the simple reason that it requires guessing a parametric guide functional form that we use to approximate a posterior.
This guide function is likely at best to be an imperfect approximation.
By paying the cost of restricting the putative posterior to have a restricted functional form, the problem of approximating a posterior is transformed to a well-posed optimization problem that seeks parameters of the putative posterior that minimize a Kullback-Leibler (KL) divergence between true posterior and the putative posterior distribution.
minimizing the KL divergence is equivalent to maximizing a criterion called the Evidence Lower Bound (ELBO), as we shall verify soon.
18.2. Prior distributions#
In order to be able to apply MCMC sampling or VI, numpyro requires that a prior distribution satisfy special properties:
we must be able to sample from it;
we must be able to compute the log pdf pointwise;
the pdf must be differentiable with respect to the parameters.
We’ll want to define a distribution class.
We will use the following priors:
a uniform distribution on \([\underline \theta, \overline \theta]\), where \(0 \leq \underline \theta < \overline \theta \leq 1\).
a truncated log-normal distribution with support on \([0,1]\) with parameters \((\mu,\sigma)\).
To implement this, let \(Z\sim N(\mu,\sigma)\) and \(\tilde{Z}\) be truncated normal with support \([-\infty,\log(1)]\), then \(\exp(Z)\) has a log normal distribution with bounded support \([0,1]\). This can be easily coded since
numpyrohas a built-in truncated normal distribution, andnumpyro’sTransformedDistributionclass that includes an exponential transformation.
a shifted von Mises distribution that has support confined to \([0,1]\) with parameter \((\mu,\kappa)\).
Let \(X\sim vonMises(0,\kappa)\). We know that \(X\) has bounded support \([-\pi, \pi]\). We can define a shifted von Mises random variable \(\tilde{X}=a+bX\) where \(a=0.5, b=1/(2 \pi)\) so that \(\tilde{X}\) is supported on \([0,1]\).
This can be implemented using
numpyro’sTransformedDistributionclass with itsAffineTransformmethod.
a truncated Laplace distribution.
We also considered a truncated Laplace distribution because its density comes in a piece-wise non-smooth form and has a distinctive spiked shape.
The truncated Laplace can be created using
numpyro’sTruncatedDistributionclass.
def truncated_log_normal_trans(loc, scale):
"""
Obtains the truncated log normal distribution
using numpyro's TruncatedNormal and ExpTransform
"""
base_dist = dist.TruncatedNormal(
low=-jnp.inf, high=jnp.log(1), loc=loc, scale=scale
)
return dist.TransformedDistribution(
base_dist, dist.transforms.ExpTransform()
)
def shifted_von_mises(κ):
"""Obtains the shifted von Mises distribution using AffineTransform"""
base_dist = dist.VonMises(0, κ)
return dist.TransformedDistribution(
base_dist,
dist.transforms.AffineTransform(loc=0.5, scale=1 / (2 * jnp.pi))
)
def truncated_laplace(loc, scale):
"""Obtains the truncated Laplace distribution on [0,1]"""
base_dist = dist.Laplace(loc, scale)
return dist.TruncatedDistribution(base_dist, low=0.0, high=1.0)
18.2.1. Variational inference#
Instead of directly sampling from the posterior, the variational inference method approximates an unknown posterior distribution with a family of tractable distributions/densities.
It then seeks to minimize a measure of statistical discrepancy between the approximating and true posteriors.
Thus variational inference (VI) approximates a posterior by solving a minimization problem.
Let the latent parameter/variable that we want to infer be \(\theta\).
Let the prior be \(p(\theta)\) and the likelihood be \(p\left(Y\vert\theta\right)\).
We want \(p\left(\theta\vert Y\right)\).
Bayes’ rule implies
where
The integral on the right side of (18.1) is typically difficult to compute.
Consider a guide distribution \(q_{\phi}(\theta)\) parameterized by \(\phi\) that we’ll use to approximate the posterior.
We choose parameters \(\phi\) of the guide distribution to minimize a Kullback-Leibler (KL) divergence between the approximate posterior \(q_{\phi}(\theta)\) and the posterior:
Thus, we want a variational distribution \(q\) that solves
Note that
For observed data \(Y\), \(p(\theta,Y)\) is a constant, so minimizing KL divergence is equivalent to maximizing
Formula (18.2) is called the evidence lower bound (ELBO).
A standard optimization routine can be used to search for the optimal \(\phi\) in our parametrized distribution \(q_{\phi}(\theta)\).
The parameterized distribution \(q_{\phi}(\theta)\) is called the variational distribution.
We can implement Stochastic Variational Inference (SVI) in numpyro using the Adam gradient descent algorithm to approximate the posterior.
We use two sets of variational distributions: Beta and TruncatedNormal with support \([0,1]\)
Learnable parameters for the Beta distribution are (\(\alpha\), \(\beta\)), both of which are positive.
Learnable parameters for the Truncated Normal distribution are (loc, scale).
Note
We restrict the truncated Normal parameter ‘loc’ to be in the interval \([0,1]\)
18.3. Implementation#
We have constructed a Python class BayesianInference that requires the following arguments to be initialized:
param: a tuple/scalar of parameters dependent on distribution typesname_dist: a string that specifies distribution names
The (param, name_dist) pair includes:
(\(\alpha\), \(\beta\), ‘beta’)
(lower_bound, upper_bound, ‘uniform’)
(loc, scale, ‘lognormal’)
Note: This is the truncated log normal.
(\(\kappa\), ‘vonMises’), where \(\kappa\) denotes concentration parameter, and center location is set to \(0.5\). Using
numpyro, this is the shifted distribution.(loc, scale, ‘laplace’)
Note: This is the truncated Laplace
The class BayesianInference has several key methods :
sample_prior:This can be used to draw a single sample from the given prior distribution.
show_prior:Plots the approximate prior distribution by repeatedly drawing samples and fitting a kernel density curve.
mcmc_sampling:INPUT: (data, num_samples, num_warmup=1000)
Takes a
jnp.arraydata and generates MCMC sampling of posterior of sizenum_samples.
svi_run:INPUT: (data, guide_dist, n_steps=10000)
guide_dist = ‘normal’ - use a truncated normal distribution as the parametrized guide
guide_dist = ‘beta’ - use a beta distribution as the parametrized guide
RETURN: (params, losses) - the learned parameters in a
dictand the vector of loss at each step.
class BayesianInference(NamedTuple):
"""
Parameters
---------
param : tuple.
a tuple object that contains all relevant parameters for the distribution
name_dist : str.
name of the distribution - 'beta', 'uniform', 'lognormal', 'vonMises', 'laplace'
rng_key : jax.random.PRNGKey
PRNG key for random number generation.
"""
param: tuple
name_dist: str
rng_key: random.PRNGKey
def create_bayesian_inference(
param: tuple,
name_dist: str,
seed: int = 0
) -> BayesianInference:
"""Factory function to create a BayesianInference instance"""
rng_key = random.PRNGKey(seed)
return BayesianInference(
param=param,
name_dist=name_dist,
rng_key=rng_key
)
def sample_prior(model: BayesianInference):
"""Define the prior distribution to sample from in numpyro models."""
if model.name_dist == "beta":
# unpack parameters
α0, β0 = model.param
sample = numpyro.sample(
"theta", dist.Beta(α0, β0), rng_key=model.rng_key
)
elif model.name_dist == "uniform":
# unpack parameters
lb, ub = model.param
sample = numpyro.sample(
"theta", dist.Uniform(lb, ub), rng_key=model.rng_key
)
elif model.name_dist == "lognormal":
# unpack parameters
loc, scale = model.param
sample = numpyro.sample(
"theta",
truncated_log_normal_trans(loc, scale),
rng_key=model.rng_key
)
elif model.name_dist == "vonMises":
# unpack parameters
κ = model.param
sample = numpyro.sample(
"theta", shifted_von_mises(κ), rng_key=model.rng_key
)
elif model.name_dist == "laplace":
# unpack parameters
loc, scale = model.param
sample = numpyro.sample(
"theta", truncated_laplace(loc, scale), rng_key=model.rng_key
)
return sample
def show_prior(
model: BayesianInference, size=1e5, bins=20, disp_plot=1
):
"""
Visualizes prior distribution by sampling from prior
and plots the approximated sampling distribution
"""
with numpyro.plate("show_prior", size=size):
sample = sample_prior(model)
# to JAX array
sample_array = jnp.asarray(sample)
# plot histogram and kernel density
if disp_plot == 1:
sns.displot(
sample_array,
kde=True,
stat="density",
bins=bins,
height=5,
aspect=1.5
)
plt.xlim(0, 1)
plt.show()
else:
return sample_array
def set_model(model: BayesianInference, data):
"""
Define the probabilistic model by specifying prior,
conditional likelihood, and data conditioning
"""
theta = sample_prior(model)
output = numpyro.sample(
"obs", dist.Binomial(len(data), theta), obs=jnp.sum(data)
)
def mcmc_sampling(
model: BayesianInference, data, num_samples, num_warmup=1000
):
"""
Computes numerically the posterior distribution
with beta prior parametrized by (α0, β0)
given data using MCMC
"""
data = jnp.array(data, dtype=float)
nuts_kernel = NUTS(set_model)
mcmc = MCMC(
nuts_kernel,
num_samples=num_samples,
num_warmup=num_warmup,
progress_bar=False,
)
mcmc.run(model.rng_key, model=model, data=data)
samples = mcmc.get_samples()["theta"]
return samples
# arguments in this function are used to align with the arguments in set_model()
# this is required by svi.run()
def beta_guide(model: BayesianInference, data):
"""
Defines the candidate parametrized variational distribution
that we train to approximate posterior with numpyro
Here we use parameterized beta
"""
α_q = numpyro.param("alpha_q", 10, constraint=constraints.positive)
β_q = numpyro.param("beta_q", 10, constraint=constraints.positive)
numpyro.sample("theta", dist.Beta(α_q, β_q))
# similar with beta_guide()
def truncnormal_guide(model: BayesianInference, data):
"""
Defines the candidate parametrized variational distribution
that we train to approximate posterior with numpyro
Here we use truncated normal on [0,1]
"""
loc = numpyro.param("loc", 0.5, constraint=constraints.interval(0.0, 1.0))
scale = numpyro.param("scale", 1, constraint=constraints.positive)
numpyro.sample(
"theta",
dist.TruncatedNormal(loc, scale, low=0.0, high=1.0)
)
def svi_init(model: BayesianInference, guide_dist, lr=0.0005):
"""Initiate SVI training mode with Adam optimizer"""
adam_params = {"lr": lr}
if guide_dist == "beta":
optimizer = Adam(step_size=lr)
svi = SVI(
set_model, beta_guide, optimizer, loss=Trace_ELBO()
)
elif guide_dist == "normal":
optimizer = Adam(step_size=lr)
svi = SVI(
set_model, truncnormal_guide, optimizer, loss=Trace_ELBO()
)
else:
print("WARNING: Please input either 'beta' or 'normal'")
svi = None
return svi
def svi_run(model: BayesianInference, data, guide_dist, n_steps=10000):
"""
Runs SVI and returns optimized parameters and losses
Returns
--------
params : the learned parameters for guide
losses : a vector of loss at each step
"""
# initiate SVI
svi = svi_init(model, guide_dist)
data = jnp.array(data, dtype=float)
result = svi.run(
model.rng_key, n_steps, model, data, progress_bar=False
)
params = dict(
(key, jnp.asarray(value)) for key, value in result.params.items()
)
losses = jnp.asarray(result.losses)
return params, losses
18.4. Alternative prior distributions#
Let’s see how well our sampling algorithm does in approximating
a log normal distribution
a uniform distribution
To examine our alternative prior distributions, we’ll plot approximate prior distributions below by calling the show_prior method.
# truncated log normal
example_ln = create_bayesian_inference(param=(0, 2), name_dist="lognormal")
show_prior(example_ln, size=100000, bins=20)
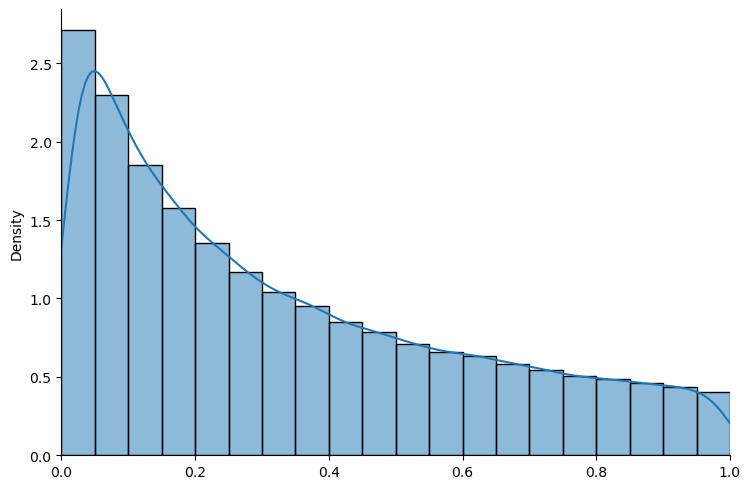
Fig. 18.1 Truncated log normal distribution#
# truncated uniform
example_un = create_bayesian_inference(param=(0.1, 0.8), name_dist="uniform")
show_prior(example_un, size=100000, bins=20)
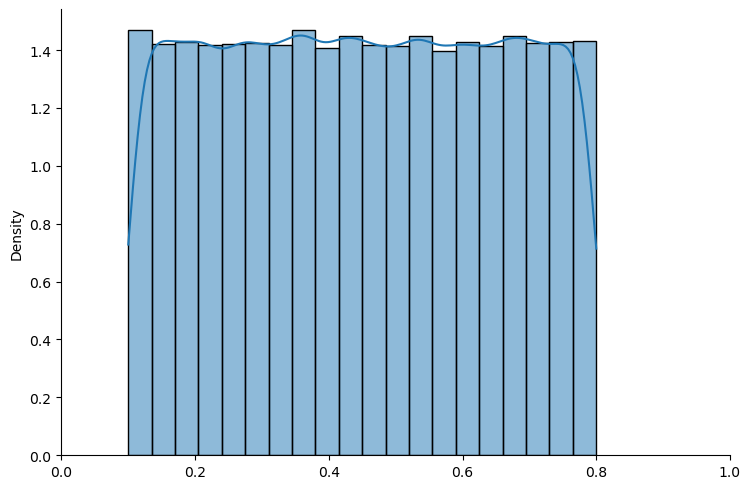
Fig. 18.2 Truncated uniform distribution#
The above graphs show that sampling seems to work well with both distributions.
Now let’s see how well things work with von Mises distributions.
# shifted von Mises
example_vm = create_bayesian_inference(param=10, name_dist="vonMises")
show_prior(example_vm, size=100000, bins=20)
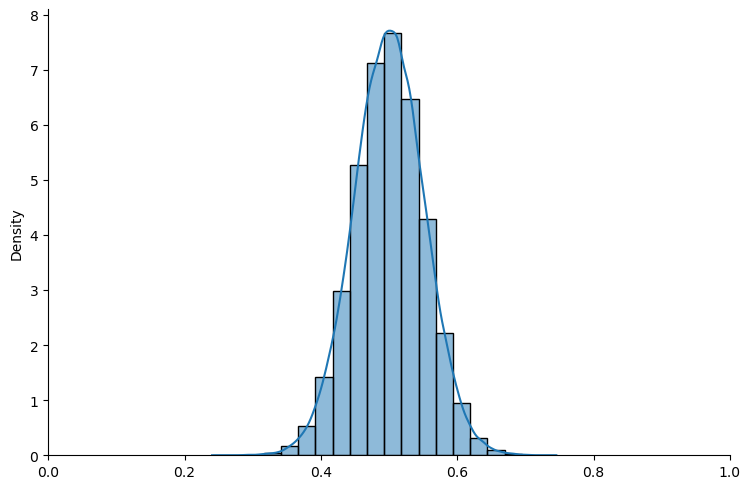
Fig. 18.3 Shifted von Mises distribution#
The graphs look good too.
Now let’s try with a Laplace distribution.
# truncated Laplace
example_lp = create_bayesian_inference(param=(0.5, 0.05), name_dist="laplace")
show_prior(example_lp, size=100000, bins=20)
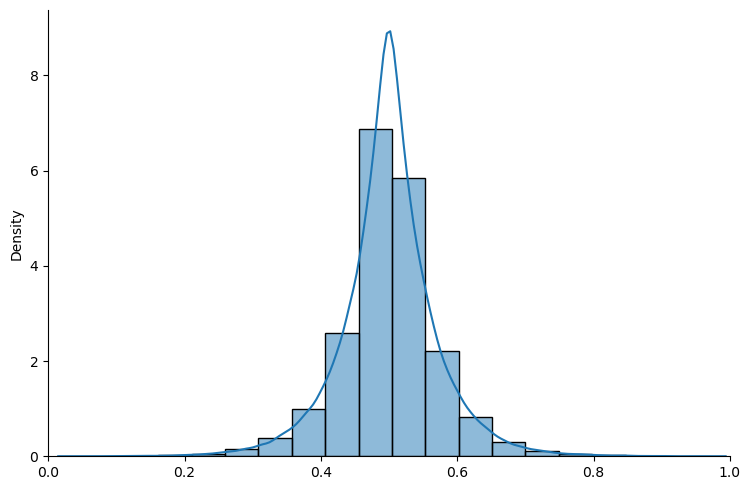
Fig. 18.4 Truncated Laplace distribution#
Having assured ourselves that our sampler seems to do a good job, let’s put it to work in using MCMC to compute posterior probabilities.
18.5. Posteriors via MCMC and VI#
We construct a class BayesianInferencePlot to implement MCMC or VI algorithms and plot multiple posteriors for different updating data sizes and different possible priors.
This class takes as inputs the true data generating parameter θ, a list of updating data sizes for multiple posterior plotting, and a defined and parametrized BayesianInference class.
It has two key methods:
BayesianInferencePlot.mcmc_plot()takes desired MCMC sample size as input and plots the output posteriors together with the prior defined inBayesianInferenceclass.BayesianInferencePlot.svi_plot()takes desired VI distribution class (‘beta’ or ‘normal’) as input and plots the posteriors together with the prior.
class BayesianInferencePlot(NamedTuple):
"""
Easily implement the MCMC and VI inference for a given instance of
BayesianInference class and plot the prior together with multiple posteriors
Parameters
----------
θ : float.
the true DGP parameter
N_list : list.
a list of sample size
bayesian_model : BayesianInference.
a class initiated using create_bayesian_inference()
binwidth : float.
plotting parameter for histogram bin width
linewidth : float.
plotting parameter for line width
colorlist : list.
list of colors for plotting
N_max : int.
maximum sample size
data : np.ndarray.
generated data array
"""
θ: float
N_list: Sequence[int]
bayesian_model: BayesianInference
binwidth: float
linewidth: float
colorlist: list
N_max: int
data: np.ndarray
def create_bayesian_inference_plot(
θ: float,
N_list: Sequence[int],
bayesian_model: BayesianInference,
*,
binwidth: float = 0.02,
linewidth: float = 0.05,
) -> BayesianInferencePlot:
"""Factory function to create a BayesianInferencePlot instance"""
colorlist = sns.color_palette(n_colors=len(N_list))
N_max = int(max(N_list))
data = simulate_draw(θ, N_max)
return BayesianInferencePlot(
θ=θ,
N_list=list(map(int, N_list)),
bayesian_model=bayesian_model,
binwidth=binwidth,
linewidth=linewidth,
colorlist=colorlist,
N_max=N_max,
data=data,
)
def mcmc_plot(
plot_model: BayesianInferencePlot, num_samples, num_warmup=1000
):
fig, ax = plt.subplots()
# plot prior
prior_sample = show_prior(
plot_model.bayesian_model, disp_plot=0
)
sns.histplot(
data=prior_sample,
kde=True,
stat="density",
binwidth=plot_model.binwidth,
color="#4C4E52",
linewidth=plot_model.linewidth,
alpha=0.1,
ax=ax,
label="Prior distribution",
)
# plot posteriors
for id, n in enumerate(plot_model.N_list):
samples = mcmc_sampling(
plot_model.bayesian_model,
plot_model.data[:n],
num_samples,
num_warmup
)
sns.histplot(
samples,
kde=True,
stat="density",
binwidth=plot_model.binwidth,
linewidth=plot_model.linewidth,
alpha=0.2,
color=plot_model.colorlist[id - 1],
label=f"Posterior with $n={n}$",
)
ax.legend(loc="upper left")
plt.xlim(0, 1)
plt.show()
def svi_fitting(guide_dist, params):
"""Fit the beta/truncnormal curve using parameters trained by SVI."""
# create x axis
xaxis = jnp.linspace(0, 1, 1000)
if guide_dist == "beta":
y = st.beta.pdf(xaxis, a=params["alpha_q"], b=params["beta_q"])
elif guide_dist == "normal":
# rescale upper/lower bound. See Scipy's truncnorm doc
lower, upper = (0, 1)
loc, scale = params["loc"], params["scale"]
a, b = (lower - loc) / scale, (upper - loc) / scale
y = st.truncnorm.pdf(
xaxis, a=a, b=b, loc=loc, scale=scale
)
return (xaxis, y)
def svi_plot(
plot_model: BayesianInferencePlot, guide_dist, n_steps=2000
):
fig, ax = plt.subplots()
# plot prior
prior_sample = show_prior(plot_model.bayesian_model, disp_plot=0)
sns.histplot(
data=prior_sample,
kde=True,
stat="density",
binwidth=plot_model.binwidth,
color="#4C4E52",
linewidth=plot_model.linewidth,
alpha=0.1,
ax=ax,
label="Prior distribution",
)
# plot posteriors
for id, n in enumerate(plot_model.N_list):
(params, losses) = svi_run(
plot_model.bayesian_model, plot_model.data[:n], guide_dist, n_steps
)
x, y = svi_fitting(guide_dist, params)
ax.plot(
x,
y,
alpha=1,
color=plot_model.colorlist[id - 1],
label=f"Posterior with $n={n}$",
)
ax.legend(loc="upper left")
plt.xlim(0, 1)
plt.show()
Let’s set some parameters that we’ll use in all of the examples below.
To save computer time at first, notice that we’ll set mcmc_num_samples = 2000 and svi_num_steps = 5000.
(Later, to increase accuracy of approximations, we’ll want to increase these.)
num_list = [5, 10, 50, 100, 1000]
mcmc_num_samples = 2000
svi_num_steps = 5000
# θ is the data generating process
true_θ = 0.8
18.5.1. Beta prior and posteriors:#
Let’s compare outcomes when we use a Beta prior.
For the same Beta prior, we shall
compute posteriors analytically
compute posteriors using MCMC using
numpyro.compute posteriors using VI using
numpyro.
Let’s start with the analytical method that we described in this Two Meanings of Probability
# first examine Beta prior
beta = create_bayesian_inference(param=(5, 5), name_dist="beta")
beta_plot = create_bayesian_inference_plot(true_θ, num_list, beta)
# plot analytical Beta prior and posteriors
xaxis = jnp.linspace(0, 1, 1000)
y_prior = st.beta.pdf(xaxis, 5, 5)
fig, ax = plt.subplots()
# plot analytical beta prior
ax.plot(xaxis, y_prior, label="Analytical Beta prior", color="#4C4E52")
data, colorlist, N_list = beta_plot.data, beta_plot.colorlist, beta_plot.N_list
# Plot analytical beta posteriors
for id, n in enumerate(N_list):
func = analytical_beta_posterior(data[:n], α0=5, β0=5)
y_posterior = func.pdf(xaxis)
ax.plot(
xaxis,
y_posterior,
color=colorlist[id - 1],
label=f"Analytical Beta posterior with $n={n}$",
)
ax.legend(loc="upper left")
plt.xlim(0, 1)
plt.show()
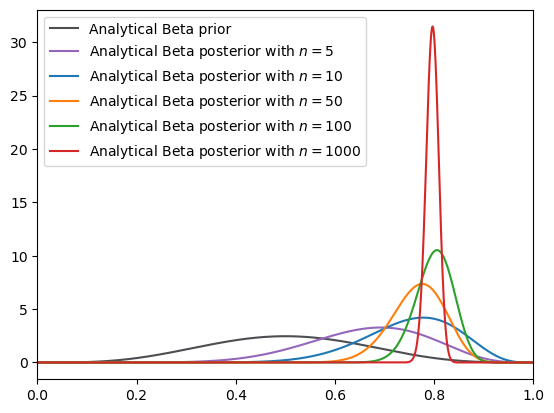
Fig. 18.5 Analytical density (Beta prior)#
Now let’s use MCMC while still using a beta prior.
We’ll do this for both MCMC and VI.
mcmc_plot(
beta_plot, num_samples=mcmc_num_samples
)
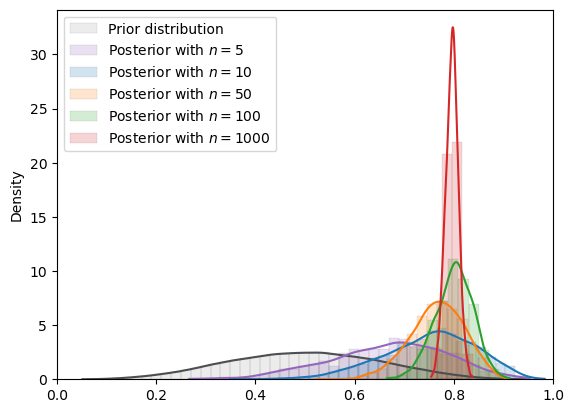
Fig. 18.6 MCMC density (Beta prior)#
svi_plot(
beta_plot, guide_dist="beta", n_steps=svi_num_steps
)
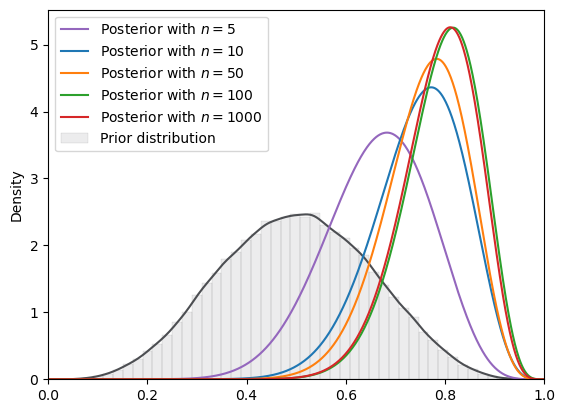
Fig. 18.7 SVI density (Beta prior, Beta guide)#
Here the MCMC approximation looks good.
But the VI approximation doesn’t look so good.
even though we use the beta distribution as our guide, the VI approximated posterior distributions do not closely resemble the posteriors that we had just computed analytically.
(Here, our initial parameter for Beta guide is (0.5, 0.5).)
But if we increase the number of steps from 5000 to 100000 in VI as we now shall do, we’ll get VI-approximated posteriors that will be more accurate, as we shall see next.
(Increasing the step size increases computational time though).
svi_plot(
beta_plot, guide_dist="beta", n_steps=100000
)
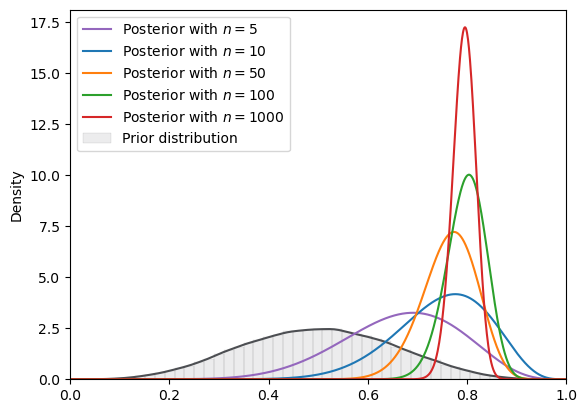
18.6. Non-conjugate prior distributions#
Having assured ourselves that our MCMC and VI methods can work well when we have a conjugate prior and so can also compute analytically, we next proceed to situations in which our prior is not a beta distribution, so we don’t have a conjugate prior.
So we will have non-conjugate priors and are cast into situations in which we can’t calculate posteriors analytically.
18.6.1. Markov chain Monte Carlo#
First, we implement and display MCMC.
We first initialize the BayesianInference classes and then can directly call BayesianInferencePlot to plot both MCMC and SVI approximating posteriors.
# Initialize BayesianInference classes
# Try uniform
std_uniform = create_bayesian_inference(param=(0, 1), name_dist="uniform")
uniform = create_bayesian_inference(param=(0.2, 0.7), name_dist="uniform")
# Try truncated log normal
lognormal = create_bayesian_inference(param=(0, 2), name_dist="lognormal")
# Try Von Mises
vonmises = create_bayesian_inference(param=10, name_dist="vonMises")
# Try Laplace
laplace = create_bayesian_inference(param=(0.5, 0.07), name_dist="laplace")
To conduct our experiments more concisely, here we define two experiment functions that will print the model information and plot the result.
def plot_mcmc_experiment(
bayesian_model: BayesianInference,
true_θ: float,
num_list: Sequence[int],
num_samples: int,
num_warmup: int = 1000,
description: str = ""
):
"""
Helper function to run and plot MCMC experiments for a given Bayesian model
"""
print(
f"=======INFO=======\n"
f"Parameters: {bayesian_model.param}\n"
f"Prior Dist: {bayesian_model.name_dist}"
)
if description:
print(description)
plot_model = create_bayesian_inference_plot(
true_θ, num_list, bayesian_model
)
mcmc_plot(plot_model, num_samples=num_samples, num_warmup=num_warmup)
def plot_svi_experiment(
bayesian_model: BayesianInference,
true_θ: float,
num_list: Sequence[int],
guide_dist: str,
n_steps: int,
description: str = ""
):
"""
Helper function to run and plot SVI experiments for a given Bayesian model
"""
print(
f"=======INFO=======\n"
f"Parameters: {bayesian_model.param}\n"
f"Prior Dist: {bayesian_model.name_dist}"
)
if description:
print(description)
plot_model = create_bayesian_inference_plot(
true_θ, num_list, bayesian_model
)
svi_plot(plot_model, guide_dist=guide_dist, n_steps=n_steps)
# Uniform
plot_mcmc_experiment(
std_uniform,
true_θ,
num_list,
mcmc_num_samples
)
=======INFO=======
Parameters: (0, 1)
Prior Dist: uniform
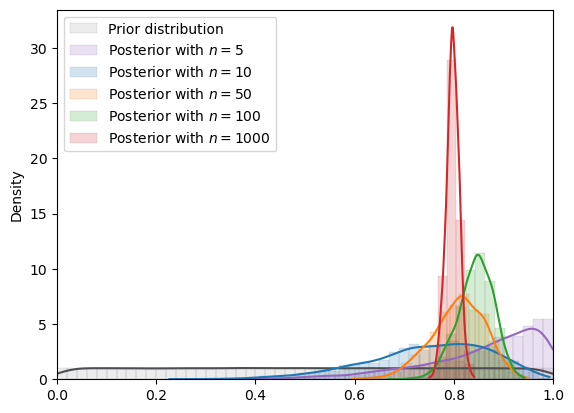
Fig. 18.8 MCMC density (uniform prior)#
plot_mcmc_experiment(
uniform,
true_θ,
num_list,
mcmc_num_samples
)
=======INFO=======
Parameters: (0.2, 0.7)
Prior Dist: uniform
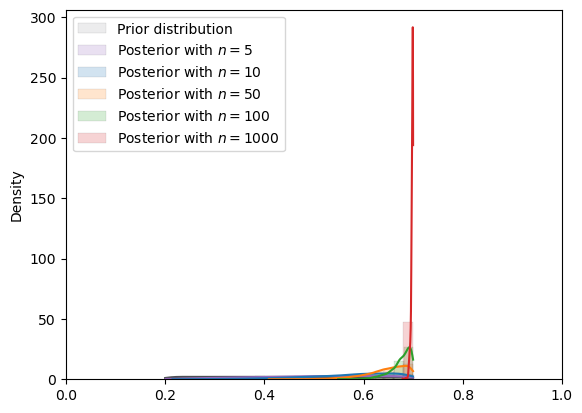
Fig. 18.9 MCMC density (uniform prior)#
In the situation depicted above, we have assumed a \(Uniform(\underline{\theta}, \overline{\theta})\) prior that puts zero probability outside a bounded support that excludes the true value.
Consequently, the posterior cannot put positive probability above \(\overline{\theta}\) or below \(\underline{\theta}\).
Note how when the true data-generating \(\theta\) is located at \(0.8\) as it is here, when \(n\) gets large, the posterior concentrates on the upper bound of the support of the prior, \(0.7\) here.
# log normal
plot_mcmc_experiment(
lognormal,
true_θ,
num_list,
mcmc_num_samples
)
=======INFO=======
Parameters: (0, 2)
Prior Dist: lognormal
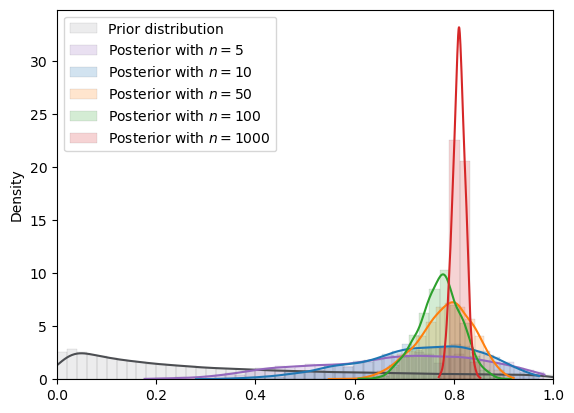
Fig. 18.10 MCMC density (log normal prior)#
# von Mises
plot_mcmc_experiment(
vonmises,
true_θ,
num_list,
mcmc_num_samples,
description="\nNOTE: Shifted von Mises"
)
=======INFO=======
Parameters: 10
Prior Dist: vonMises
NOTE: Shifted von Mises
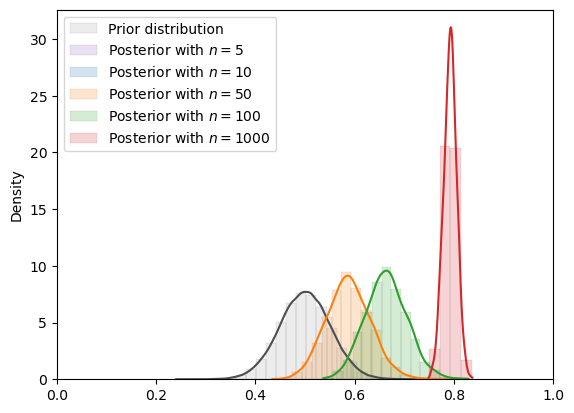
Fig. 18.11 MCMC density (von Mises prior)#
# Laplace
plot_mcmc_experiment(
laplace,
true_θ,
num_list,
mcmc_num_samples
)
=======INFO=======
Parameters: (0.5, 0.07)
Prior Dist: laplace
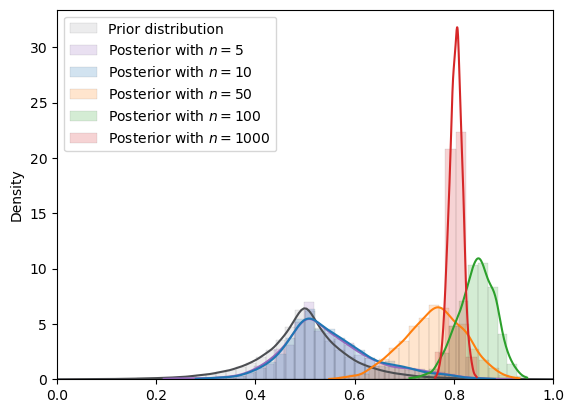
Fig. 18.12 MCMC density (Laplace prior)#
18.6.2. Variational inference#
To get more accuracy we will now increase the number of steps for Variational Inference (VI)
svi_num_steps = 50000
18.6.2.1. VI with a truncated normal guide#
# Uniform
plot_svi_experiment(
create_bayesian_inference(param=(0, 1), name_dist="uniform"),
true_θ,
num_list,
"normal",
svi_num_steps
)
=======INFO=======
Parameters: (0, 1)
Prior Dist: uniform
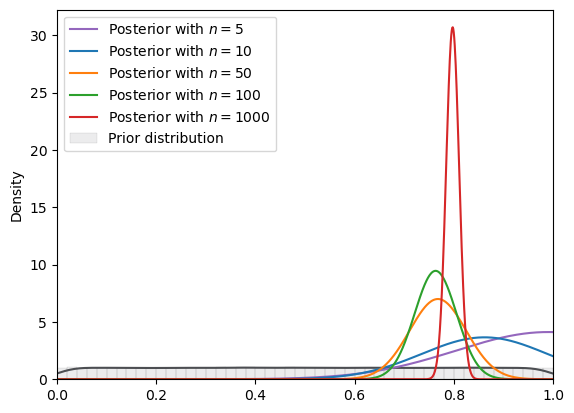
Fig. 18.13 SVI density (uniform prior, normal guide)#
# log normal
plot_svi_experiment(
lognormal,
true_θ,
num_list,
"normal",
svi_num_steps
)
=======INFO=======
Parameters: (0, 2)
Prior Dist: lognormal
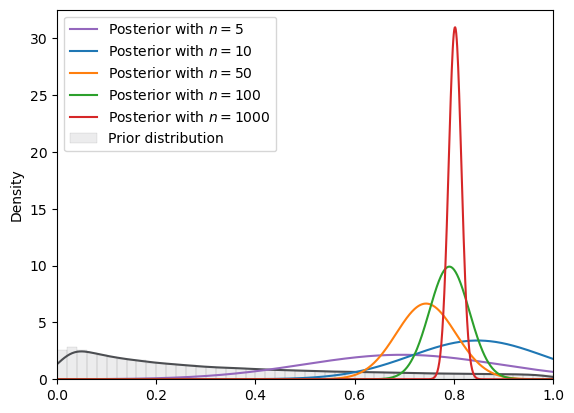
Fig. 18.14 SVI density (log normal prior, normal guide)#
# Laplace
plot_svi_experiment(
laplace,
true_θ,
num_list,
"normal",
svi_num_steps
)
=======INFO=======
Parameters: (0.5, 0.07)
Prior Dist: laplace
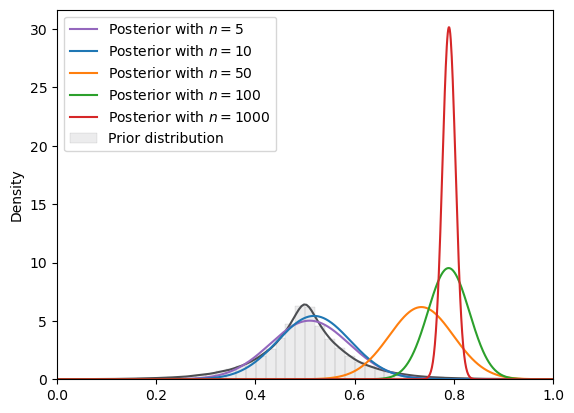
Fig. 18.15 SVI density (Laplace prior, normal guide)#
18.6.2.2. Variational inference with a Beta guide distribution#
# uniform
plot_svi_experiment(
std_uniform,
true_θ,
num_list,
"beta",
svi_num_steps
)
=======INFO=======
Parameters: (0, 1)
Prior Dist: uniform
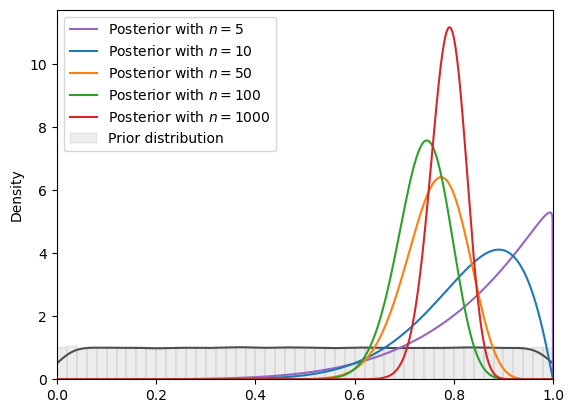
Fig. 18.16 SVI density (uniform prior, Beta guide)#
# log normal
plot_svi_experiment(
lognormal,
true_θ,
num_list,
"beta",
svi_num_steps
)
=======INFO=======
Parameters: (0, 2)
Prior Dist: lognormal
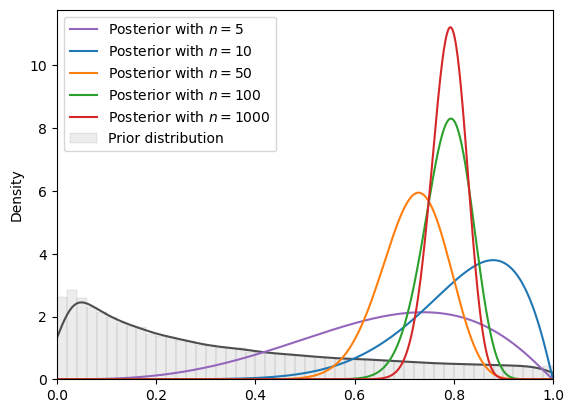
Fig. 18.17 SVI density (log normal prior, Beta guide)#
# von Mises
plot_svi_experiment(
vonmises,
true_θ,
num_list,
"beta",
svi_num_steps,
description="Shifted von Mises"
)
=======INFO=======
Parameters: 10
Prior Dist: vonMises
Shifted von Mises
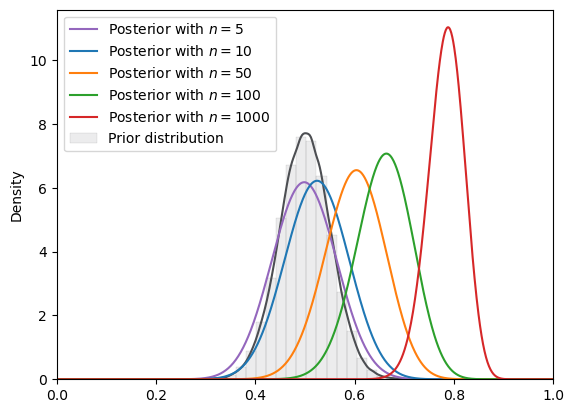
Fig. 18.18 SVI density (von Mises prior, Beta guide)#
# Laplace
plot_svi_experiment(
laplace,
true_θ,
num_list,
"beta",
svi_num_steps
)
=======INFO=======
Parameters: (0.5, 0.07)
Prior Dist: laplace
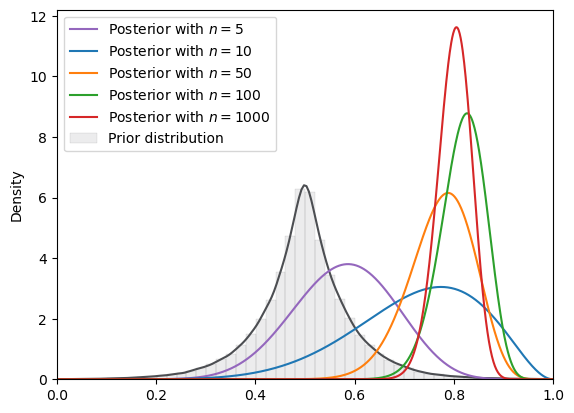
Fig. 18.19 SVI density (Laplace prior, Beta guide)#