20. Forecasting an AR(1) Process#
!pip install arviz pymc
This lecture describes methods for forecasting statistics that are functions of future values of a univariate autogressive process.
The methods are designed to take into account two possible sources of uncertainty about these statistics:
random shocks that impinge of the transition law
uncertainty about the parameter values of the AR(1) process
We consider two sorts of statistics:
prospective values \(y_{t+j}\) of a random process \(\{y_t\}\) that is governed by the AR(1) process
sample path properties that are defined as non-linear functions of future values \(\{y_{t+j}\}_{j \geq 1}\) at time \(t\)
Sample path properties are things like “time to next turning point” or “time to next recession”.
To investigate sample path properties we’ll use a simulation procedure recommended by Wecker [Wecker, 1979].
To acknowledge uncertainty about parameters, we’ll deploy pymc to construct a Bayesian joint posterior distribution for unknown parameters.
Let’s start with some imports.
import numpy as np
import arviz as az
import pymc as pmc
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style('white')
colors = sns.color_palette()
import logging
logging.basicConfig()
logger = logging.getLogger('pymc')
logger.setLevel(logging.CRITICAL)
20.1. A Univariate First-Order Autoregressive Process#
Consider the univariate AR(1) model:
where the scalars \(\rho\) and \(\sigma\) satisfy \(|\rho| < 1\) and \(\sigma > 0\); \(\{\epsilon_{t+1}\}\) is a sequence of i.i.d. normal random variables with mean \(0\) and variance \(1\).
The initial condition \(y_{0}\) is a known number.
Equation (20.1) implies that for \(t \geq 0\), the conditional density of \(y_{t+1}\) is
Further, equation (20.1) also implies that for \(t \geq 0\), the conditional density of \(y_{t+j}\) for \(j \geq 1\) is
The predictive distribution (20.3) that assumes that the parameters \(\rho, \sigma\) are known, which we express by conditioning on them.
We also want to compute a predictive distribution that does not condition on \(\rho,\sigma\) but instead takes account of our uncertainty about them.
We form this predictive distribution by integrating (20.3) with respect to a joint posterior distribution \(\pi_t(\rho,\sigma | y^t)\) that conditions on an observed history \(y^t = \{y_s\}_{s=0}^t\):
Predictive distribution (20.3) assumes that parameters \((\rho,\sigma)\) are known.
Predictive distribution (20.4) assumes that parameters \((\rho,\sigma)\) are uncertain, but have known probability distribution \(\pi_t(\rho,\sigma | y^t )\).
We also want to compute some predictive distributions of “sample path statistics” that might include, for example
the time until the next “recession”,
the minimum value of \(Y\) over the next 8 periods,
“severe recession”, and
the time until the next turning point (positive or negative).
To accomplish that for situations in which we are uncertain about parameter values, we shall extend Wecker’s [Wecker, 1979] approach in the following way.
first simulate an initial path of length \(T_0\);
for a given prior, draw a sample of size \(N\) from the posterior joint distribution of parameters \(\left(\rho,\sigma\right)\) after observing the initial path;
for each draw \(n=0,1,...,N\), simulate a “future path” of length \(T_1\) with parameters \(\left(\rho_n,\sigma_n\right)\) and compute our three “sample path statistics”;
finally, plot the desired statistics from the \(N\) samples as an empirical distribution.
20.2. Implementation#
First, we’ll simulate a sample path from which to launch our forecasts.
In addition to plotting the sample path, under the assumption that the true parameter values are known, we’ll plot \(.9\) and \(.95\) coverage intervals using conditional distribution (20.3) described above.
We’ll also plot a bunch of samples of sequences of future values and watch where they fall relative to the coverage interval.
def AR1_simulate(rho, sigma, y0, T):
# Allocate space and draw epsilons
y = np.empty(T)
eps = np.random.normal(0, sigma, T)
# Initial condition and step forward
y[0] = y0
for t in range(1, T):
y[t] = rho * y[t-1] + eps[t]
return y
def plot_initial_path(initial_path):
"""
Plot the initial path and the preceding predictive densities
"""
# Compute .9 confidence interval]
y0 = initial_path[-1]
center = np.array([rho**j * y0 for j in range(T1)])
vars = np.array([sigma**2 * (1 - rho**(2 * j)) / (1 - rho**2) for j in range(T1)])
y_bounds1_c95, y_bounds2_c95 = center + 1.96 * np.sqrt(vars), center - 1.96 * np.sqrt(vars)
y_bounds1_c90, y_bounds2_c90 = center + 1.65 * np.sqrt(vars), center - 1.65 * np.sqrt(vars)
# Plot
fig, ax = plt.subplots(1, 1, figsize=(12, 6))
ax.set_title("Initial Path and Predictive Densities", fontsize=15)
ax.plot(np.arange(-T0 + 1, 1), initial_path)
ax.set_xlim([-T0, T1])
ax.axvline(0, linestyle='--', alpha=.4, color='k', lw=1)
# Simulate future paths
for i in range(10):
y_future = AR1_simulate(rho, sigma, y0, T1)
ax.plot(np.arange(T1), y_future, color='grey', alpha=.5)
# Plot 90% CI
ax.fill_between(np.arange(T1), y_bounds1_c95, y_bounds2_c95, alpha=.3, label='95% CI')
ax.fill_between(np.arange(T1), y_bounds1_c90, y_bounds2_c90, alpha=.35, label='90% CI')
ax.plot(np.arange(T1), center, color='red', alpha=.7, label='expected mean')
ax.legend(fontsize=12)
plt.show()
sigma = 1
rho = 0.9
T0, T1 = 100, 100
y0 = 10
# Simulate
np.random.seed(145)
initial_path = AR1_simulate(rho, sigma, y0, T0)
# Plot
plot_initial_path(initial_path)
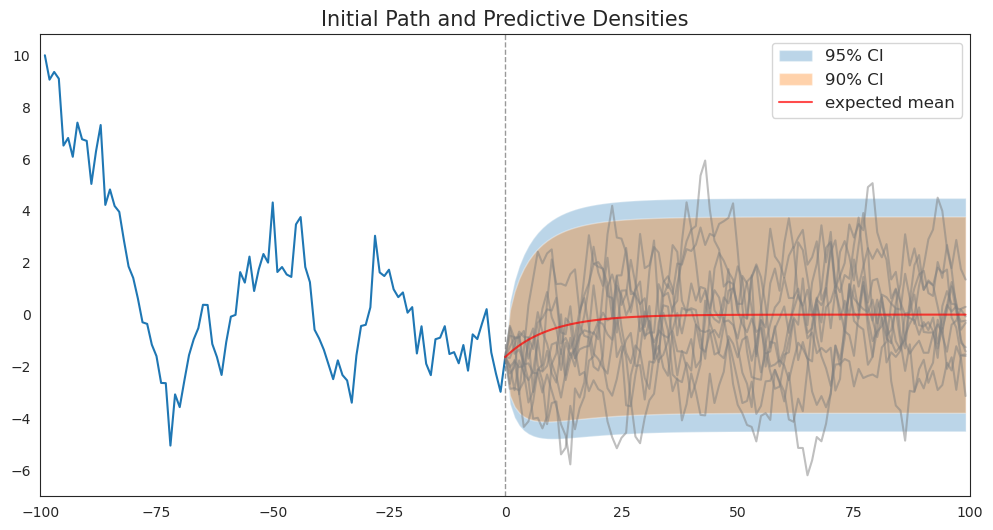
As functions of forecast horizon, the coverage intervals have shapes like those described in https://python.quantecon.org/perm_income_cons.html
20.3. Predictive Distributions of Path Properties#
Wecker [Wecker, 1979] proposed using simulation techniques to characterize predictive distribution of some statistics that are non-linear functions of \(y\).
He called these functions “path properties” to contrast them with properties of single data points.
He studied two special prospective path properties of a given series \(\{y_t\}\).
The first was time until the next turning point.
he defined a “turning point” to be the date of the second of two successive declines in \(y\).
To examine this statistic, let \(Z\) be an indicator process
Then the random variable time until the next turning point is defined as the following stopping time with respect to \(Z\):
Wecker [Wecker, 1979] also studied the minimum value of \(Y\) over the next 8 quarters which can be defined as the random variable.
It is interesting to study yet another possible concept of a turning point.
Thus, let
Define a positive turning point today or tomorrow statistic as
This is designed to express the event
“after one or two decrease(s), \(Y\) will grow for two consecutive quarters”
Following [Wecker, 1979], we can use simulations to calculate probabilities of \(P_t\) and \(N_t\) for each period \(t\).
20.4. A Wecker-Like Algorithm#
The procedure consists of the following steps:
index a sample path by \(\omega_i\)
for a given date \(t\), simulate \(I\) sample paths of length \(N\)
for each path \(\omega_i\), compute the associated value of \(W_t(\omega_i), W_{t+1}(\omega_i), \dots\)
consider the sets \(\{W_t(\omega_i)\}^{T}_{i=1}, \ \{W_{t+1}(\omega_i)\}^{T}_{i=1}, \ \dots, \ \{W_{t+N}(\omega_i)\}^{T}_{i=1}\) as samples from the predictive distributions \(f(W_{t+1} \mid \mathcal y_t, \dots)\), \(f(W_{t+2} \mid y_t, y_{t-1}, \dots)\), \(\dots\), \(f(W_{t+N} \mid y_t, y_{t-1}, \dots)\).
20.5. Using Simulations to Approximate a Posterior Distribution#
The next code cells use pymc to compute the time \(t\) posterior distribution of \(\rho, \sigma\).
Note that in defining the likelihood function, we choose to condition on the initial value \(y_0\).
def draw_from_posterior(sample):
"""
Draw a sample of size N from the posterior distribution.
"""
AR1_model = pmc.Model()
with AR1_model:
# Start with priors
rho = pmc.Uniform('rho',lower=-1.,upper=1.) # Assume stable rho
sigma = pmc.HalfNormal('sigma', sigma = np.sqrt(10))
# Expected value of y at the next period (rho * y)
yhat = rho * sample[:-1]
# Likelihood of the actual realization.
y_like = pmc.Normal('y_obs', mu=yhat, sigma=sigma, observed=sample[1:])
with AR1_model:
trace = pmc.sample(10000, tune=5000)
# check condition
with AR1_model:
az.plot_trace(trace, figsize=(17, 6))
rhos = trace.posterior.rho.values.flatten()
sigmas = trace.posterior.sigma.values.flatten()
post_sample = {
'rho': rhos,
'sigma': sigmas
}
return post_sample
post_samples = draw_from_posterior(initial_path)
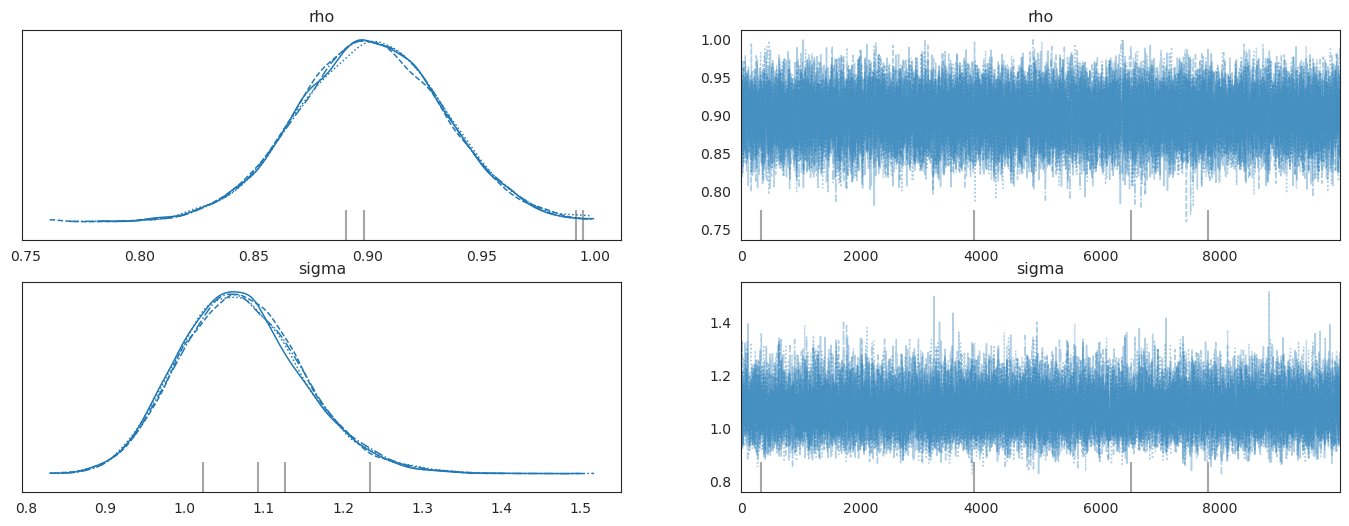
The graphs on the left portray posterior marginal distributions.
20.6. Calculating Sample Path Statistics#
Our next step is to prepare Python code to compute our sample path statistics.
# define statistics
def next_recession(omega):
n = omega.shape[0] - 3
z = np.zeros(n, dtype=int)
for i in range(n):
z[i] = int(omega[i] <= omega[i+1] and omega[i+1] > omega[i+2] and omega[i+2] > omega[i+3])
if np.any(z) == False:
return 500
else:
return np.where(z==1)[0][0] + 1
def minimum_value(omega):
return min(omega[:8])
def severe_recession(omega):
z = np.diff(omega)
n = z.shape[0]
sr = (z < -.02).astype(int)
indices = np.where(sr == 1)[0]
if len(indices) == 0:
return T1
else:
return indices[0] + 1
def next_turning_point(omega):
"""
Suppose that omega is of length 6
y_{t-2}, y_{t-1}, y_{t}, y_{t+1}, y_{t+2}, y_{t+3}
that is sufficient for determining the value of P/N
"""
n = np.asarray(omega).shape[0] - 4
T = np.zeros(n, dtype=int)
for i in range(n):
if ((omega[i] > omega[i+1]) and (omega[i+1] > omega[i+2]) and
(omega[i+2] < omega[i+3]) and (omega[i+3] < omega[i+4])):
T[i] = 1
elif ((omega[i] < omega[i+1]) and (omega[i+1] < omega[i+2]) and
(omega[i+2] > omega[i+3]) and (omega[i+3] > omega[i+4])):
T[i] = -1
up_turn = np.where(T == 1)[0][0] + 1 if (1 in T) == True else T1
down_turn = np.where(T == -1)[0][0] + 1 if (-1 in T) == True else T1
return up_turn, down_turn
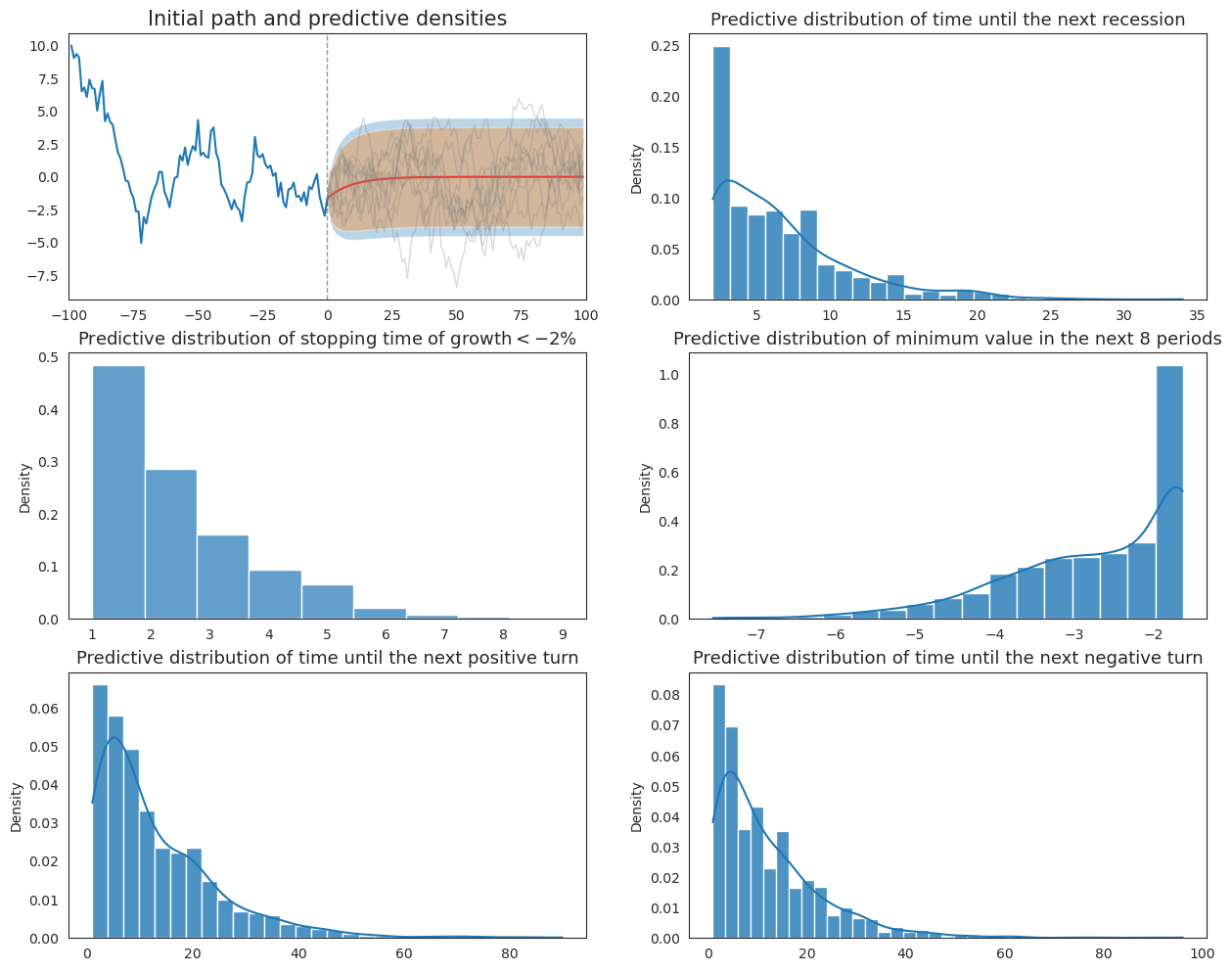
20.7. Original Wecker Method#
Now we apply Wecker’s original method by simulating future paths and compute predictive distributions, conditioning on the true parameters associated with the data-generating model.
def plot_Wecker(initial_path, N, ax):
"""
Plot the predictive distributions from "pure" Wecker's method.
"""
# Store outcomes
next_reces = np.zeros(N)
severe_rec = np.zeros(N)
min_vals = np.zeros(N)
next_up_turn, next_down_turn = np.zeros(N), np.zeros(N)
# Compute .9 confidence interval]
y0 = initial_path[-1]
center = np.array([rho**j * y0 for j in range(T1)])
vars = np.array([sigma**2 * (1 - rho**(2 * j)) / (1 - rho**2) for j in range(T1)])
y_bounds1_c95, y_bounds2_c95 = center + 1.96 * np.sqrt(vars), center - 1.96 * np.sqrt(vars)
y_bounds1_c90, y_bounds2_c90 = center + 1.65 * np.sqrt(vars), center - 1.65 * np.sqrt(vars)
# Plot
ax[0, 0].set_title("Initial path and predictive densities", fontsize=15)
ax[0, 0].plot(np.arange(-T0 + 1, 1), initial_path)
ax[0, 0].set_xlim([-T0, T1])
ax[0, 0].axvline(0, linestyle='--', alpha=.4, color='k', lw=1)
# Plot 90% CI
ax[0, 0].fill_between(np.arange(T1), y_bounds1_c95, y_bounds2_c95, alpha=.3)
ax[0, 0].fill_between(np.arange(T1), y_bounds1_c90, y_bounds2_c90, alpha=.35)
ax[0, 0].plot(np.arange(T1), center, color='red', alpha=.7)
# Simulate future paths
for n in range(N):
sim_path = AR1_simulate(rho, sigma, initial_path[-1], T1)
next_reces[n] = next_recession(np.hstack([initial_path[-3:-1], sim_path]))
severe_rec[n] = severe_recession(sim_path)
min_vals[n] = minimum_value(sim_path)
next_up_turn[n], next_down_turn[n] = next_turning_point(sim_path)
if n%(N/10) == 0:
ax[0, 0].plot(np.arange(T1), sim_path, color='gray', alpha=.3, lw=1)
# Return next_up_turn, next_down_turn
sns.histplot(next_reces, kde=True, stat='density', ax=ax[0, 1], alpha=.8, label='True parameters')
ax[0, 1].set_title("Predictive distribution of time until the next recession", fontsize=13)
sns.histplot(severe_rec, kde=False, stat='density', ax=ax[1, 0], binwidth=0.9, alpha=.7, label='True parameters')
ax[1, 0].set_title(r"Predictive distribution of stopping time of growth$<-2\%$", fontsize=13)
sns.histplot(min_vals, kde=True, stat='density', ax=ax[1, 1], alpha=.8, label='True parameters')
ax[1, 1].set_title("Predictive distribution of minimum value in the next 8 periods", fontsize=13)
sns.histplot(next_up_turn, kde=True, stat='density', ax=ax[2, 0], alpha=.8, label='True parameters')
ax[2, 0].set_title("Predictive distribution of time until the next positive turn", fontsize=13)
sns.histplot(next_down_turn, kde=True, stat='density', ax=ax[2, 1], alpha=.8, label='True parameters')
ax[2, 1].set_title("Predictive distribution of time until the next negative turn", fontsize=13)
fig, ax = plt.subplots(3, 2, figsize=(15,12))
plot_Wecker(initial_path, 1000, ax)
plt.show()
20.8. Extended Wecker Method#
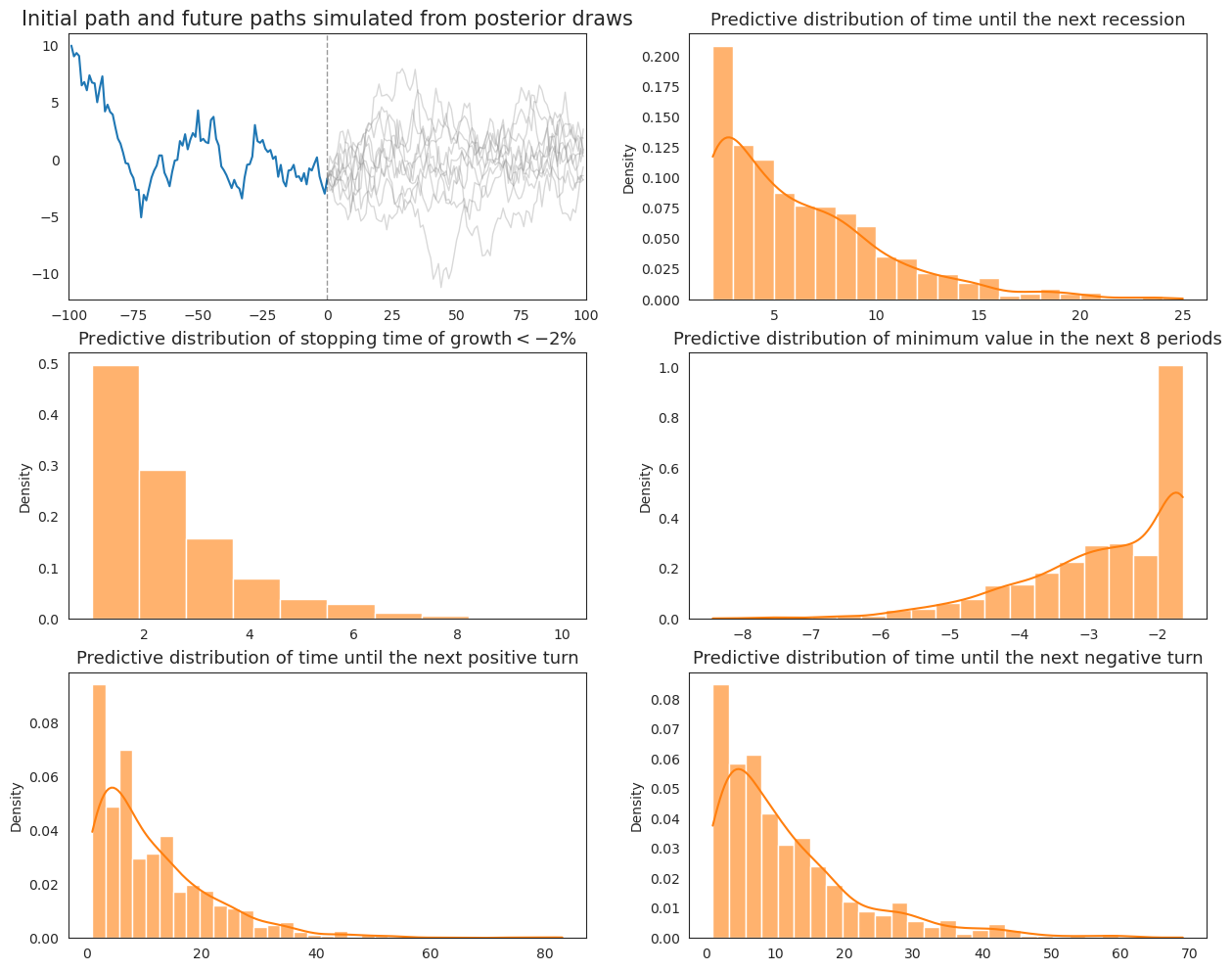
Now we apply we apply our “extended” Wecker method based on predictive densities of \(y\) defined by (20.4) that acknowledge posterior uncertainty in the parameters \(\rho, \sigma\).
To approximate the intergration on the right side of (20.4), we repeatedly draw parameters from the joint posterior distribution each time we simulate a sequence of future values from model (20.1).
def plot_extended_Wecker(post_samples, initial_path, N, ax):
"""
Plot the extended Wecker's predictive distribution
"""
# Select a sample
index = np.random.choice(np.arange(len(post_samples['rho'])), N + 1, replace=False)
rho_sample = post_samples['rho'][index]
sigma_sample = post_samples['sigma'][index]
# Store outcomes
next_reces = np.zeros(N)
severe_rec = np.zeros(N)
min_vals = np.zeros(N)
next_up_turn, next_down_turn = np.zeros(N), np.zeros(N)
# Plot
ax[0, 0].set_title("Initial path and future paths simulated from posterior draws", fontsize=15)
ax[0, 0].plot(np.arange(-T0 + 1, 1), initial_path)
ax[0, 0].set_xlim([-T0, T1])
ax[0, 0].axvline(0, linestyle='--', alpha=.4, color='k', lw=1)
# Simulate future paths
for n in range(N):
sim_path = AR1_simulate(rho_sample[n], sigma_sample[n], initial_path[-1], T1)
next_reces[n] = next_recession(np.hstack([initial_path[-3:-1], sim_path]))
severe_rec[n] = severe_recession(sim_path)
min_vals[n] = minimum_value(sim_path)
next_up_turn[n], next_down_turn[n] = next_turning_point(sim_path)
if n % (N / 10) == 0:
ax[0, 0].plot(np.arange(T1), sim_path, color='gray', alpha=.3, lw=1)
# Return next_up_turn, next_down_turn
sns.histplot(next_reces, kde=True, stat='density', ax=ax[0, 1], alpha=.6, color=colors[1], label='Sampling from posterior')
ax[0, 1].set_title("Predictive distribution of time until the next recession", fontsize=13)
sns.histplot(severe_rec, kde=False, stat='density', ax=ax[1, 0], binwidth=.9, alpha=.6, color=colors[1], label='Sampling from posterior')
ax[1, 0].set_title(r"Predictive distribution of stopping time of growth$<-2\%$", fontsize=13)
sns.histplot(min_vals, kde=True, stat='density', ax=ax[1, 1], alpha=.6, color=colors[1], label='Sampling from posterior')
ax[1, 1].set_title("Predictive distribution of minimum value in the next 8 periods", fontsize=13)
sns.histplot(next_up_turn, kde=True, stat='density', ax=ax[2, 0], alpha=.6, color=colors[1], label='Sampling from posterior')
ax[2, 0].set_title("Predictive distribution of time until the next positive turn", fontsize=13)
sns.histplot(next_down_turn, kde=True, stat='density', ax=ax[2, 1], alpha=.6, color=colors[1], label='Sampling from posterior')
ax[2, 1].set_title("Predictive distribution of time until the next negative turn", fontsize=13)
fig, ax = plt.subplots(3, 2, figsize=(15, 12))
plot_extended_Wecker(post_samples, initial_path, 1000, ax)
plt.show()
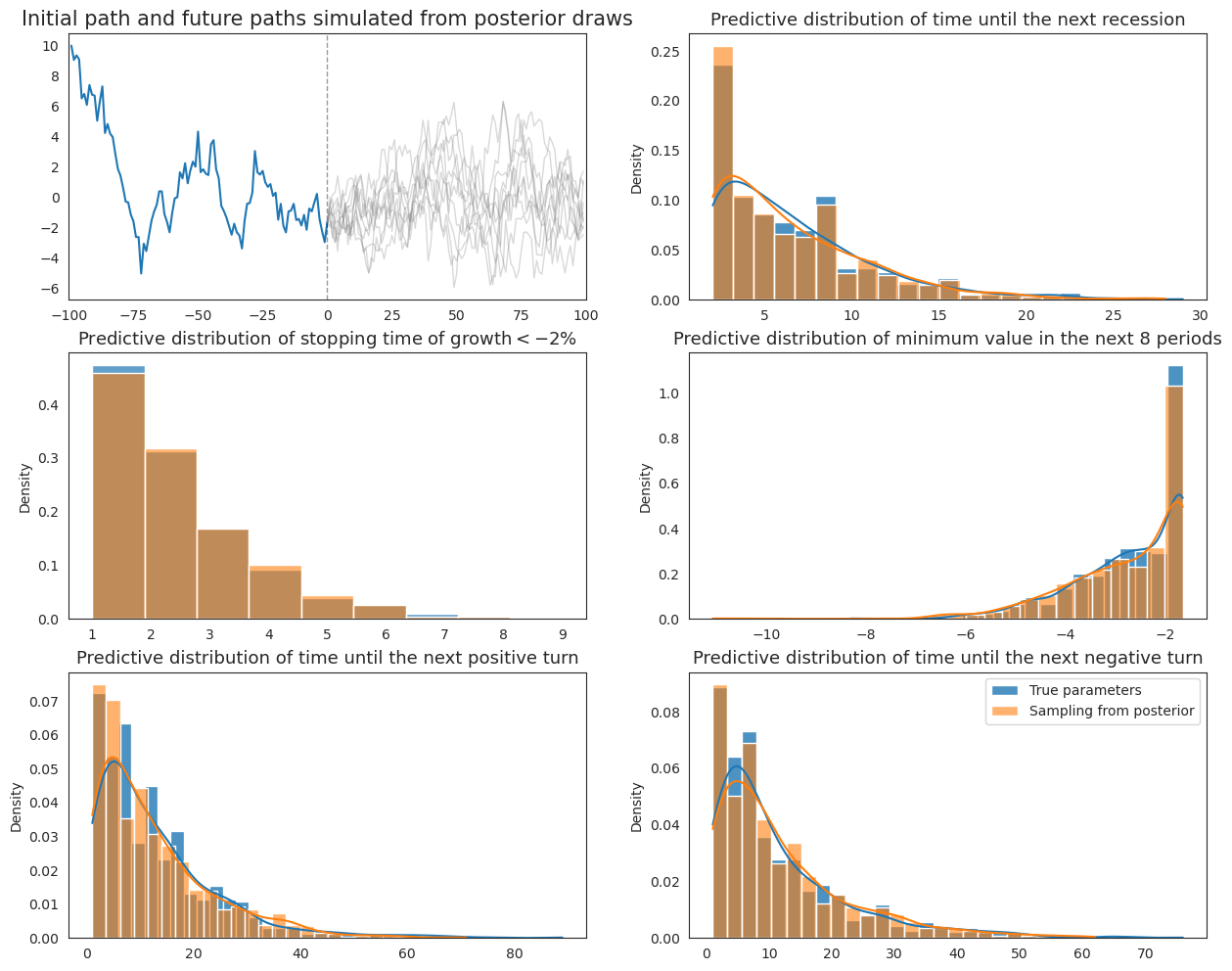
20.9. Comparison#
Finally, we plot both the original Wecker method and the extended method with parameter values drawn from the posterior together to compare the differences that emerge from pretending to know parameter values when they are actually uncertain.
fig, ax = plt.subplots(3, 2, figsize=(15,12))
plot_Wecker(initial_path, 1000, ax)
ax[0, 0].clear()
plot_extended_Wecker(post_samples, initial_path, 1000, ax)
plt.legend()
plt.show()